Astroturf. It's Real, Real Fake Grass: Part 1
Jan 25, 2023
6 mins read

The real motive of this series of blogs may take a few iterations to become clear to the reader, assuming of course I can actually pull it off. Let’s forget about the big picture for a moment and break things down into pieces I can wrap my head around. Mostly because I’ve already started, I’m already doing the things, and I don’t even know the right words to call them.
Disclaimer: I’m gonna use words that sound right to me, in no way am I guaranteeing that they are the correct (mathematical?) verbiage or terminology.
For the purposes of this article, we’ll focus on the objective at hand: Loading and navigating the network graph of english language Wikipedia.
Network Graphs
I learned about network graphs and graph theory because there was this story about this dude with some bridges and some questions.
At a high level, the url you are viewing right now https://remyhax.xyz/posts/astroturf/ can be thought of as a node. The Wikipedia page about the Seven Bridges of Königsberg can also be thought of as a node.
The node of the current page and the node of the Seven Bridges of Königsberg article share a relationship (the link seen above) hereby called an edge.
If you click the link above and navigate to the Wikiped article, you will not find any links that lead back to the current page you are reading. The relationship, or edge, has direction.

The above example is very tightly scoped. As you can probably imagine, there are many other links that lead to the page you’re currently reading. Let’s add 2.

As you can probably imagine, there is also a lot of links to other Wikipedia articles from within the “Seven Bridges” article. Let’s add some from just the first section of the article.
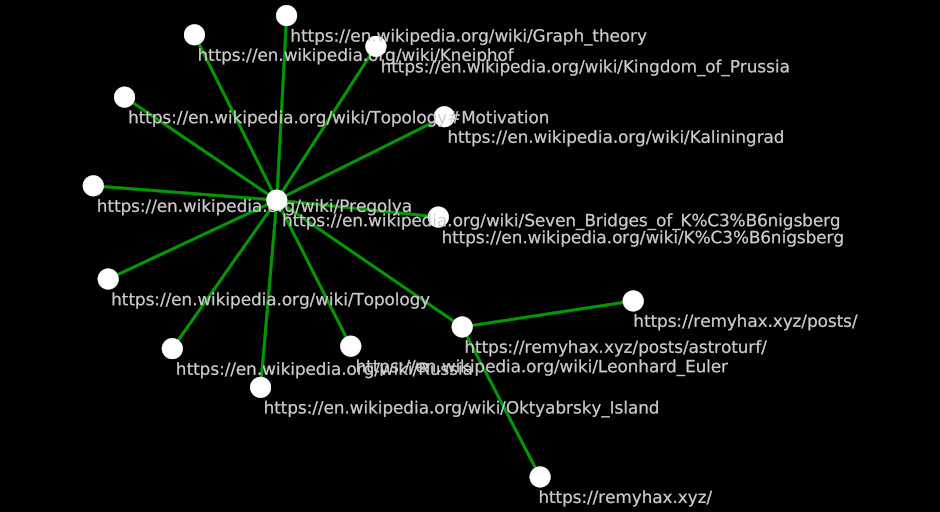
As you can see, things can get very complicated very quickly.
Loading Wikipedia
Wikipedia provides dumps of their entire archive in various formats here: https://meta.wikimedia.org/wiki/Data_dump_torrents#English_Wikipedia
For my example I will be using enwiki-20211020-pages-articles-multistream.xml which clocks in at ~85GB uncompressed.
A thing that I really like about XML is that you can easily iterate over it using tokens and don’t need to load the entire thing into memory. After writing a small module to insert rows into a SQLite database representing source-->destination edges between wikipedia pages, I’m able to write some code that parses the large XML file and writes the flattened rows to a database we can later query and manipulate.
package main
import (
"bufio"
"flag"
"fmt"
"net/url"
"os"
"strings"
"github.com/dustin/go-wikiparse"
"github.com/schollz/progressbar/v3"
"github.com/xen0bit/wikinetwork/pkg/dbtools"
)
var inputFile = flag.String("infile", "enwiki-latest-pages-articles.xml", "Input file path")
type Redirect struct {
Title string `xml:"title,attr"`
}
type Page struct {
Title string `xml:"title"`
Redir Redirect `xml:"redirect"`
Text string `xml:"revision>text"`
}
func Canonicalize(text string) string {
can := strings.ToLower(text)
can = strings.Replace(can, " ", "_", -1)
can = url.QueryEscape(can)
return can
}
func main() {
flag.Parse()
xmlFile, err := os.Open(*inputFile)
if err != nil {
fmt.Println("Error opening file:", err)
return
}
defer xmlFile.Close()
fio := bufio.NewReader(xmlFile)
p, err := wikiparse.NewParser(fio)
if err != nil {
panic(err)
}
sc := dbtools.NewSqliteConn("test.db")
if sc.Err != nil {
panic(err)
}
if err := sc.CreateTables(); err != nil {
panic(err)
}
s := make(chan struct{})
rc := make(chan *dbtools.Edge)
go sc.InsertEdges(rc, s)
bar := progressbar.Default(-1)
for err == nil {
var page *wikiparse.Page
page, err = p.Next()
if err == nil && page.Redir.Title == "" {
ct := Canonicalize(page.Title)
l := wikiparse.FindLinks(page.Revisions[0].Text)
for _, v := range l {
rc <- &dbtools.Edge{
Source: ct,
Destination: Canonicalize(v),
}
bar.Add(1)
}
}
}
bar.Finish()
close(rc)
<-s
sc.DB.Close()
}
While the code may not be the most efficient, it was sufficient.
According to my logging, the code wrote 352,663,589 edges to the database in 1h32m15s (63715 rows/sec). Which was enough time for me to write the blog to this very line you’re reading.
Analysis
Now that we have a database of our edges, we can perform some queries to get some hints about the nature of the network.
First a sanity check that all the reported edges made it into the database.
sqlite> select count(*) from edges;
352663589
Cool this matches the reported logs. Let’s take a quick peek at the first 10 rows.
sqlite> select * from edges limit 10;
anarchism|political_philosophy
anarchism|political_movement
anarchism|authority
anarchism|hierarchy
anarchism|state_%28polity%29
anarchism|left-wing
anarchism|political_spectrum
anarchism|libertarian_marxism
anarchism|libertarian
anarchism|libertarian_socialism
Nice, we see that as expected, we’ve inserted rows representing edges from the source “anarchism” to the destination’s of various page titles.
As part of our Go code above, we ignored pages where redirects were included, such as (3-methyl-2-oxobutanoate dehydrogenase (acetyl-transferring)) kinase which make the graph significantly more sparse. Let’s check for articles which are “dead ends”, or articles which are linked to, but do not have any outgoing links back to another top level page. Kinda like if you drove into a neighborhood and hit a cul-de-sac.
sqlite> select count(distinct(destination)) from edges
where destination not in (
select distinct(source) from edges
);
24429111
Nice, so there’s ~24.5M page destination’s we can remove to make sure that our network only focuses on densely interconnected nodes. As there will inevitably be multiple source pages pointing to these “dead end”’s, we’ll be deleting more than 25.5M edges.
sqlite> delete from edges
where destination not in (
select distinct(source) from edges
);
After removing these stragglers, we are left with
sqlite> select count(*) from edges;
257454584
…edges.
257,454,584 edges is a lot. But it’s still 95,209,005 less than what we started with.
Most network graphing software I’ve worked with claims to support somewhere in the range of ~1M edges/nodes, so we’ll do a quick visualization with a smaller truncated list (500k), then move on to the nitty gritty.
sqlite> .headers on
sqlite> .mode csv
sqlite> .output network.csv
sqlite> select * from edges limit 500000;
sqlite> .quit
Quickly throwing this into https://cosmograph.app/ gives us a nice visulization of this 500k subsection of our graph.
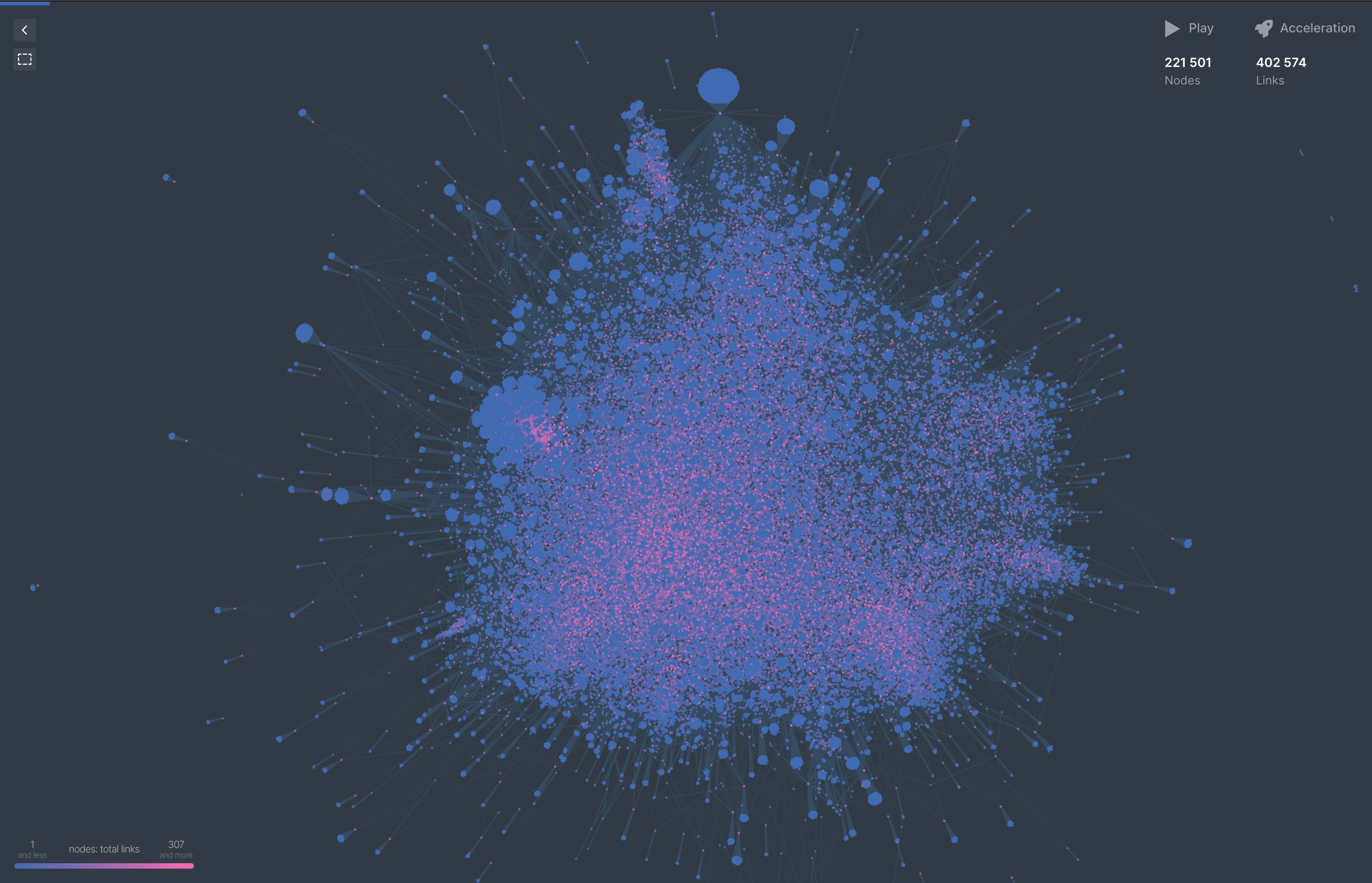
The total stats we remain to work with is:
| Nodes | Edges |
|---|---|
| 9,593,395 | 257,454,584 |
Until next time.
Sharing is caring!